Accelerating the Development and Uptake of Rapid Diagnostics to Address Antibiotic Resistance: Proceedings of a Workshop (2023)
Chapter: 2 Exploring the Need for Rapid Diagnostics
2
Exploring the Need for Rapid Diagnostics
Karen C. Carroll (Director, Division Medical Microbiology and Professor of Pathology, Johns Hopkins University School of Medicine) provided an overview of the current state of rapid diagnostic development and gaps that rapid diagnostics might be best suited to address (e.g., to support targeted treatment decisions or to enable real-time surveillance). Workshop speakers
then discussed how rapid diagnostics fit into the healthcare system and how “success” in this area could be defined.
Antimicrobial resistance (AMR) is ranked by the WHO as one of the top ten global public health threats facing humanity (WHO, 2021). By 2050, it is projected that AMR will be responsible for 10 million deaths and a loss of $100 trillion to the global economy due to loss of productivity (O’Neill, 2016). In the United States alone, there are 2.8 million antibiotic-resistant infections each year, resulting in 35,000 deaths. Further, some of the most urgent threats to health, according to Carroll, include multi-drug resistant gram-negative bacteria (e.g., carbapenem-resistant Acinetobacter, Enterobacterales), drug-resistant Neisseria gonorrhoeae (NG), Candida auris, and Clostridioides difficile (C. diff) (CDC, 2019a). The coronavirus disease 2019 (COVID-19) pandemic has exacerbated the global AMR crisis, she added.
Patient Perspective
“I was the kid who invented his own cure for a nonhealing wound caused by a relentless, antibiotic-resistant infection,” said Bradley Burnam (AMR survivor and Founder, Turn Therapeutics). While working in hospitals as a device representative, Burnam contracted an antimicrobial-resistant skin infection, which resulted in months of pain, surgeries, and antibiotic treatment. He ultimately developed his own ointment that put an end to the recurring infections. However, the “romantic tales of garage laboratories” do not tell the full story, said Burnam. The stories did not adequately describe the fear of seeing half of his face purple with a double-in-size cellulitic ear or mention the infectious disease specialist who advised emergency surgery and a powerful course of intravenous (IV) antibiotics. “They don’t talk about the sound of the scalpel cutting, scraping” for hours, or “describe the smell of the electrocautery” from burning tissue, he said. The stories do not mention that no anesthesiologist was available to numb his senses, or that it took nearly two hours to affix 50 stitches to piece his face back together. They did not speak to the months of antibiotic treatment and the effects on his gut or the four months he spent with deep abscesses in his face and skull that were packed with gauze and topical antibiotics. These “antiquated tools offered nothing but allergic responses and occasional comfort,” said Burnam.
Sadly, he said, the story of his hospital-acquired infection is not uncommon; thousands of patients suffer from drug-resistant infections each year. A future in which the supply of antibiotics no longer keeps pace with the organisms is a real and current scenario: humans did not administer the first dose of a systemic antibiotic until 1941, and it took microbes less than a decade to defeat it (Dhingra et al., 2020).
As a patient, said Burnam, he is angry that in the last 20 years there have been only four new antibiotic class drugs approved. At the current pace of drug approval, resistant mutations could develop before a new antibiotic is approved. According to Burnam, it may be time to take dramatic steps. For example, antibiotic resistance could be declared a national health emergency, opening pathways for Emergency Use Authorization (EUA) of drugs and devices. Legislation could be passed that would incentivize investors to deploy capital and support the economic viability of AMR products. Antibiotics are becoming rare as a class of therapy, said Burnam, but the diseases they treat are common. Burnam suggests that investors and developers “love orphan drugs” because of the lower hurdles to completing clinical trials, high rate of reimbursement, and longer exclusivity periods. In contrast, antibiotics do not benefit from federal incentives such as EUA, expedited review, transferable exclusivity vouchers, orphan drug designations, tax incentives, and reimbursement subsidies.
Bioethics Perspective
Treatment decisions involving antibiotics—including whether to prescribe an antibiotic, which antibiotic to use, and the appropriate duration of use—are incorrect in 30 to 50 percent of cases, said Tracey L. Cohen (Distinguished Visiting Scholar, Institute for Bioethics & Health Policy, University of Miami Miller School of Medicine) (Ventola, 2015). When clinicians are unable to rapidly identify a causative pathogen, this may lead to the following scenarios:
- a patient is prescribed an antibiotic that is unwarranted (e.g., for a viral infection);
- a patient is prescribed an incorrect antibiotic;
- a patient is prescribed a powerful, broad-spectrum antibiotic to cover the widest range of possible bacteria;
- a patient is denied antibiotics until test results are available;
- a patient is denied antibiotics altogether.
In each of these scenarios, said Cohen, healthcare providers are harming their patients. Giving the wrong antibiotic or broad-spectrum antibiotics increase the risk of resistance within the patient, which increases the likelihood of treatment failure. Delaying or denying treatment can lead to deterioration of the condition and/or dangerous complications (e.g., bronchitis leading to pneumonia, strep leading to heart damage). The harms that result from improper antibiotic decision-making violate the bioethical duties that healthcare providers owe to their patients, said Cohen. These duties include the responsibility of beneficence (to do good, to heal) and
non-maleficence (to do no harm). In addition, the healthcare provider’s duty to respect the patient’s autonomy is infringed. Cohen explained that patients are owed the right to full informed consent, which includes the risks and benefits of a proposed treatment and of any alternatives. However, when providers are lacking information about pathogens, patients are not given accurate information about risks, benefits, and alternatives. This results in patients relying on treatment plans to their detriment, emphasized Cohen.
Rapid point-of-care diagnostics, said Cohen, can help ensure that treatment decisions are made based on empirical data rather than supposition. They could assist providers in upholding their bioethical duties to patients, while also helping to combat the antibiotic resistance crisis. Cohen pointed out that pharmaceutical and biotechnology companies also have an obligation of non-maleficence; they should ensure that rapid diagnostics do not sacrifice accuracy for speed because inaccurate results can lead to harmful treatment decisions.
New diagnostics may be significantly more expensive than traditional methods, a distinction that requires bioethical and equity considerations among several different parties, according to Cohen. Healthcare providers and institutions must offer life-saving diagnostics to all patients, regardless of their ability to pay. Pharmaceutical and biotechnology companies should find ways to keep costs low to promote accessibility. State and federal governments have an ethical obligation to help forestall the antibiotic resistance crisis to ensure the health of citizens. These stakeholders should work towards subsidizing rapid tests (as happened with COVID-19) to ensure they are available to all. Further, governments should ensure that diagnostics are widely distributed and available, particularly among healthcare providers working in low-income communities. Third party payers have an ethical obligation to cover the costs of these diagnostics. Cohen emphasized that rapid diagnostics should be regarded as routine tests rather than specialty tests, which can be excluded from coverage, and that failure of insurers to pay would be an unjustifiable interference with healthcare providers’ ethical obligations to patients. She observed that it is more cost-effective for payers to pay for these tests because they will cut long-term costs by reducing morbidity and mortality.
In summary, said Cohen, rapid diagnostics will help healthcare providers mitigate improper treatment decisions which harm patients in violation of the bioethical principles. Stakeholders—including pharmaceutical and biotechnology companies, government entities, healthcare providers, and third-party payers—have corresponding bioethical imperatives to create, distribute, utilize, and subsidize reliable, rapid point-of-care diagnostics. However, Cohen emphasized that diagnostics must be justly distributed and financially accessible in conformity with the principle of social justice. As the WHO has observed, the antibiotic resistance crisis may seem slow-moving and abstract
compared to the COVID-19 pandemic. However, antibiotic resistance is a growing concern for everyone around the globe, and the problem demands a collective focused attention.
Clinical Microbiology Laboratory Perspective
Clinical microbiology laboratories largely depend on traditional culture-based techniques (e.g., inoculation of specimens to plated media). There are advantages to these techniques, Carroll said: they are cost-effective, they have undergone extensive clinical validation, and microbiologists are comfortable performing them. However, traditional techniques tend to be “very slow” (Miller et al., 2019). Newer methods have been developed, said Carroll, but these tend to be “add-on” approaches rather than a replacement for traditional diagnostic tests, in part due to limitations in the spectrum of pathogens that can be detected, variable specificity and sensitivity, lack of differentiation between living and dead cells, and cost. Further culture-based methods can test for multiple pathogens simultaneously, which may be particularly useful when there is no a priori knowledge of the causative pathogen.
According to Carroll, phenotypic susceptibility testing continues to be important in the detection of AMR. Antimicrobial susceptibility testing (AST) relies on semi-automated and automated devices that use microbroth dilution methods. This approach is standardized and quantitative, and there are interpretive guidelines to determine susceptibility versus resistance. However, Carroll emphasized that this approach is slow, not amenable for testing all bacterial pathogens, and there is a lag between the availability of new agents and incorporation into AST panels by manufacturers. Clinical and Laboratory Standards Institute (CLSI)1 and the European Committee on Antimicrobial Susceptibility Testing (EUCAST)2 are continually reevaluating the guidelines for susceptibility and resistance, said Carroll, which makes it challenging to incorporate new criteria for interpreting AST panels in a timely manner. A survey found that up to 70 percent of clinical laboratories in the United States are not using the current CLSI criteria for determination of resistance (Simner et al., 2022). This is a patient safety issue, she said; quality AST is critical for infection control, hospital and national surveillance, and antibiograms for empiric treatment.
Carroll noted that recent advances show some promise. For example, the Accelerate Pheno is a Food and Drug Administration (FDA)-approved platform that can identify organisms in positive blood cultures within one
___________________
1 See https://clsi.org/standards/?page=1&sort=date&sortdir=desc&subcat=AST&area= (accessed January 25, 2023).
2 See https://www.eucast.org/ast_of_bacteria (accessed January 25, 2023).
hour using fluorescence in-situ hybridization probes and can detect phenotypic susceptibility within seven hours using microscopic image analysis (Marschal et al., 2017). This approach allows clinicians to rapidly tailor treatment within a reasonably short period of time (8 hours) and can improve antimicrobial stewardship. However, there are technical barriers for some “drug/bug” combinations that add time and cost to the process, and there is financial risk to the industry in bringing these to the market. There are other new approaches as well, said Carroll, such as deoxyribonucleic acid (DNA) amplification methods that target specific genes, immunochromatographic assays, and antibiotic degradations assays. These approaches can detect resistance very rapidly (within 15 to 60 minutes) and confirm phenotypic resistance, which improves antimicrobial stewardship and infection control. However, Carroll highlighted some limitations to these approaches. For example, a negative result does not imply susceptibility, a positive resistance marker does not necessarily confer phenotypic resistance, testing may be limited to certain antibiotics, and there are additional associated costs.
Over the last several decades, disruptive technologies have advanced diagnostics in clinical labs and made transformative changes. Some culture-based methods have been replaced by multiplexed molecular syndromic panel tests, and organisms can be identified faster using Matrix Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS). MALDI-TOF MS identifies organisms based on their unique protein profile and requires only a single colony rather than a large biomass. This approach is cost-effective, highly accurate, and can be used for bacteria, fungi, and mycobacteria. It has been shown to have a “tremendous medical impact,” said Carroll, including reducing time to therapy and length of stay for patients (Miller et al., 2019). Beyond identification, mass spectrometry can be used for susceptibility testing and strain typing for epidemiology. Another notable advance has been the development of syndromic panel tests, which combine organism detection and resistance marker detection for bacterial pathogens. These can be used to detect the major causes of a particular syndrome such as respiratory infections, gastroenteritis, or meningitis. These tests have been particularly useful, said Carroll, in reducing the turnaround time for antiviral and antibacterial therapy. However, there are limited studies on patient outcomes, with most studies showing variable impact on outcomes such as length of stay and mortality (Ramanan et al., 2017). Carroll emphasized the need for guidelines for appropriate utilization of syndromic panel tests. Other transformative methods on the horizon include DNA-microarray based hybridization technology, T2MR technology3, rapid phenotypic/genotypic
___________________
3 See https://www.t2biosystems.com/products-technology/t2mr-technology/
susceptibility testing, total laboratory automation combined with artificial intelligence (AI), and next generation sequencing.
According to Carroll, next generation sequencing (NGS) is the “next paradigm shift” in diagnostics. There are many applications of NGS, including whole genome sequencing, targeted amplification, and metagenomic NGS (mNGS). Whole genome sequencing is already being used in public health labs for a variety of purposes, including identifying the resistance genes in an organism, and tracking outbreaks of foodborne illness. The real power of this technology, however, is in mNGS, said Carroll, where the genomes within a clinical specimen are subjected to massive parallel sequencing and sophisticated data analysis to arrive at a diagnosis. In traditional culture-based testing, a large proportion of samples are negative for a pathogen due to pretesting treatment, uncultivatable pathogens, or the need for special handling. In contrast, mNGS is effective at detecting a broad range of pathogens. However, Carroll noted that there are multiple disadvantages of mNGS. It is expensive (around $2500 per sample), it is complicated and has no reliable reference method, turnaround time is not fast (around 5 days on average), and it requires a huge investment in laboratory infrastructure. As a result, according to Carroll, mNGS is currently used as a “method of last resort.” To overcome these challenges, Carroll highlighted several potential solutions, including utilizing existing molecular workflows, implementing newer technologies to speed up actionable results, and developing universal well-standardized metrics (Table 2-1).
In summary, Carroll said that despite technological advances in AMR testing in clinical microbiology, there remain several challenges. Issues including workforce shortages, lack of institutional investment in clinical laboratories, and regulatory impediments are hindering the development and implementation of new tools that could transform testing and the care of patients. Further, she noted a need for diagnostic stewardship and the implementation of guidelines to help ensure appropriate test utilization and optimize patient care.
Point-of-care diagnostics
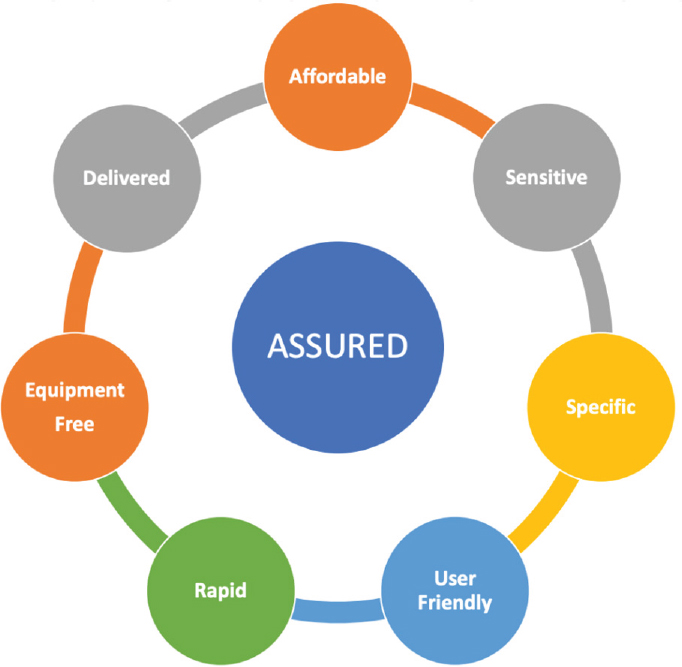
There are a number of exciting new tools in the point-of-care (POC) testing toolbox, said Carroll. The WHO developed criteria for POC tests: they should be affordable, sensitive, specific, user-friendly, rapid, equipment-free, and deliverable to where the patient is located (ASSURED) (Figure 2-1).4 While these criteria were developed with sexually transmitted infections in mind, she said, they can be applied to all rapid diagnostics. In
___________________
4 See https://apps.who.int/iris/bitstream/handle/10665/68990/TDR_STI_IDE_04.1.pdf (accessed January 25, 2023).
TABLE 2-1 Challenges to the Use of mNGS and Potential Solutions
| Current Challenges | Potential Solutions |
|---|---|
Requires investment in laboratory infrastructure
|
Use of existing molecular workflows
|
| Costs | Use of commercially available systems Limit use to diagnostic dilemmas Prospective cost-effectiveness studies |
Not amenable to immediate to fast TAT (5 days on average)
|
|
| Complicated validation (Laboratory developed tests, no reliable reference method) | Use of published protocols More universal well-standardized metrics needed |
NOTE: POC = point-of-care; TAT = turnaround time
SOURCE: Presented by Karen Carroll, October 13, 2022.
2019, Land et al. (2019) added two more criteria—Real-time connectivity and Ease of specimen collection—making the acronym REASSURED.
Emerging POC testing technologies include microfluidics, biosensors, digital droplet polymerase chain reaction (PCR), and paper-based devices. Carroll noted that these technologies show promise; for example, microfluidics combined with isothermal amplification allows for fast thermal cycling and high sensitivity at the POC. However, there is a need for more research on the barriers to realizing the REASSURED criteria, specifically, the cost of translating these technologies into the clinical environment. During the COVID-19 pandemic, diagnostic development accelerated: clinical labs created lab-developed tests, labs partnered with industry to fast-track technologies, and the government provided research support and regulatory flexibility. Carroll shared several examples of microfluidics-based technologies that were used for POC testing for COVID-19 (Table 2-2).
The promise of POC testing is most evident for certain conditions, said Carroll, including sexually transmitted infections (STIs), tuberculosis, urinary tract infections (UTIs), respiratory infections, malaria, and neglected tropical diseases. There are more than 2.4 million cases of
SOURCE: Presented by Karen Carroll, October 13, 2022.
syphilis, gonorrhea, and chlamydia in the United States each year, and resistance among Neisseria gonorrhoeae isolates is high (CDC, 2019b). Other STIs of interest include emerging Mycoplasma genitalium, human papilloma virus, Trichomonas vaginalis and herpes simplex 1 and 2. There are several current and future POC tests for STIs; most provide results within 30 minutes and are comparable to the high-throughput instruments currently in clinical labs. Carroll emphasized that the short timeframe is critical because studies show that patients presenting to STI clinics or the emergency department (ED) do not want to “hang around” for more than 45 minutes; the ability to see patients, diagnose them, and treat them within a 45-minute window is “very powerful.” A workshop participant added that there is a need to create a system for simultaneous testing for STIs and resistance markers, rather than conducting them serially, due to these time constraints.
TABLE 2-2 Examples of Microfluidics-Based Tests Developed for SARS-CoV-2
| POC Test | Assay Chemistry | Sample Type | Fluid Activation/Control | Signal Detection | Connectivity |
|---|---|---|---|---|---|
|
Cue Health https://cuehealth.com/ |
Isothermal | Nasal swab | Capillary/wax valves Fluid mixing by sonication | Electrochemical | Portable Bluetooth connected reader/mobile app |
| Visby Medical https://www.visbymedical.com/ | RT-PCR | Nasal swab | Gear motor/rotary on chip valves | Colorimetric (LFA) | None |
|
Abbott ID Now https://www.abbott.com/ |
Isothermal | Nasal/NP/throat | Manual/Manual | Fluorescence | Portable instrument with LED screen |
NOTE: LCD = liquid-crystal display; LFA = lateral flow assay; NP = nasopharyngeal;
POC = point-of-care; RT-PCR = reverse transcription-polymerase chain reaction
SOURCE: Presented by Karen Carroll, October 13, 2022.
Prior to the pandemic, studies demonstrated the effectiveness of tests using self-collected samples (e.g., I Want the Kit, I Know, and TakeMeHome). In combination with telemedicine, STI clinics turned to home collection tests in the early days of the pandemic. Carroll highlighted the benefits and challenges associated with self-collection tests. They are convenient, private, cost-effective, accurate, and readily accepted by patients; these benefits lead to increased testing and thus increased detection and treatment of STIs. The challenges of self-collection tests include language or literacy barriers for some patients (including those most at need), low specimen return rates, difficulty with surveillance and contact tracing, regulatory issues, and concerns about false positives or negatives.
In summary, said Carroll, there have been unprecedented technological advances that hold promise for enhanced diagnostics in labs and at the point-of-care, but that there are a fair number of hurdles that need to be addressed to optimize implementation. These hurdles include
- technical barriers and costs to mass production/commercialization;
- need for better outcome studies to understand patient impact;
- need for studies to understand workflow barriers in clinics and laboratories;
- quality management considerations (e.g., contamination, poor user performance);
- data management;
- regulatory and reimbursement issues for laboratories and industry.
Developer Perspective
The test development process, said Craig Whiteford (Senior Director R&D, Becton Dickinson) starts with the basic question: “What is the need?” Based on the answer to this question, design requirements are developed to describe how the test will fulfill the need. In the case of AMR, he said, the need is for rapid antimicrobial testing for susceptibility or resistance, and design requirements include
- time to results;
- target (e.g., bacteria);
- specimen (e.g., whole blood, respiratory specimen);
- impact on clinical workflow (e.g., does a new test require additional time and staff?);
- regulatory considerations;
- cost.
There are almost always tradeoffs among these design details, said Whiteford: for example, a test may be able to deliver results very quickly but be cost-prohibitive. Developers must also decide on the appropriate technological approach; in the case of AMR, will a test use a phenotypic, genetic, or genomic approach? Phenotypic approaches are limited due to cell division, which takes time and cannot be sped up. Genetic approaches (e.g., PCR) work well and are fairly quick, said Whiteford, but are limited in terms of targets. Genomic approaches (massive parallel sequencing) are the “great promise” of testing in this area.
Whiteford shared a Pugh Matrix that illustrates the benefits and drawbacks of different types of AST and AMR testing (Table 2-3). The first two columns show traditional manual or automated tests (e.g., disk diffusion) which have many advantages but they take more time. The third column shows the limitations of rapid traditional tests (e.g., microfluidic phenotypic systems) in terms of targets, specimen, costs, and clinical workflow. Whiteford explained that these tests are adjunct to other testing, putting a burden on the laboratory and the cost. Genomic predictive tests, seen in the fourth column, represent a new approach that holds great promise; however, these tests are currently limited due to reliance on AI. Whiteford emphasized a need to build and share a database of information for AI to work with, but there is no current regulatory guidance on AI. Moreover, there is a need for further discussion and collaboration among stakeholders for approaches that use AI, and, additionally, genomic predictive tests are currently quite expensive.
Shown on the right-hand side of the matrix, genetic AMR tests (e.g., PCR) can produce results within 8 hours, but they are limited in terms of targets and the impact on workflow. These tests usually only provide information on resistance markers, which tell a provider what not to treat with but not what should be used. The pros and cons of the genomic approach for AMR testing are similar to the genomic approach for AST. There are also technological barriers to both AST and AMR tests, he said, including the “needle in the haystack” issue: if there are only a few colony-forming units (CFUs) per milliliter of specimen, it can be difficult to ensure that the CFUs are detected by the test.
Reimbursement Perspective
Susan Van Meter (President, American Clinical Laboratory Association) indicated that during the COVID-19 pandemic, the importance of leveraging every modality for testing became clear. Developing and utilizing tests for a variety of settings (e.g., inpatient, ambulatory, consumer) reduced barriers to access and supported clinicians in their workflow. Similarly, Van Meter emphasized a need to think broadly about the kinds of tests that will be useful for antimicrobial resistance. There is tremendous value
TABLE 2-3 Pugh Matrix of AST and AMR Testing Approaches
| Requirements | AST | AMR | |||||
|---|---|---|---|---|---|---|---|
| Trad. Manual* | Trad. Automated | Rapid Trad. | Genomic Predictive | Genetic PCR/Iso | Genomic | ||
| Targeted | Metagenomic | ||||||
| Time to Results (≤8 hrs.) | x | x | ✓ | Library Prep | ✓ | Library Prep | Library Prep |
| Target(s) | ✓ | ✓ | Limited | ✓ | Limited | ✓ | ✓ |
| Specimen(s) | ✓ | ✓ | Limited | Limited | ✓ | Limited | Limited |
| Clinical Workflow | ✓ | ✓ | Adjunct | SA | Adjunct | SA | SA |
| IVDD/IVDR | Gold Std | ✓ | ✓ | AI | ✓ | AI | |
| Cost | $ | $ | $$ | $$$ | $$ | $$$ | $$$ |
| Comprehensive | ✓ Available | * Baseline | SA = Stand Alone | |
| Limited / Potential | ||||
| X / $$$ |
NOTE: AI = artificial intelligence; AMR = antimicrobial resistance; AST = antimicrobial susceptibility testing; Iso = isothermal; IVDD = In Vitro Diagnostic Medical Devices Directive; IVDR = In Vitro Diagnostic Medical Devices Regulation; PCR = polymerase chain reaction; Std = standard; Trad = traditional
SOURCE: Presented by Craig Whitford, October 13, 2022.
in diagnostic tests that help ensure the right patient gets the right treatment at the right time; effective diagnostics save time and effort for both patients and providers and ultimately reduce costs. However, the value of a test is not always easily translated into an appropriate reimbursement. Van Meter added that oversight of the development to tests can cause uncertainty for developers, explaining that there is a lack of an overarching regulatory apparatus for diagnostic tests and technologies. Some tests are developed in laboratories; these laboratory developed tests (LDTs) are regulated by the Clinical Laboratory improvement Amendments (CLIA) under the Centers for Medicare & Medicaid Services (CMS). Other tests are developed for the commercial market and regulated by the FDA. There is some discussion in Congress about modernizing the regulation of diagnostics to create a single risk-based framework, but Van Meter said she does not think the chances of this becoming law are particularly high.
When it comes to bringing a test to market, Van Meter noted that developers need to consider whether there is an existing Current Procedural Terminology (CPT) code for the test. Applying to the American Medical Association for a new code can take between six months and two years. Payer coverage for tests can be complicated and may vary among geographic regions. For example, contractors who provide services to Medicare patients may make local coverage decisions and coverage may vary depending on whether the patient is in an inpatient or ambulatory setting. Within hospitals, coverage for a test is generally rolled into an inpatient Diagnosis Related Group (DRG) rather than reimbursed separately. Coverage in the ambulatory setting is more complicated. If a physician’s office provides a test onsite, they will generally bill the payer directly, whereas if samples are collected and sent to a commercial lab, the lab will generally bill the payer. Van Meter noted that Medicare reimbursement amounts for clinical laboratory fees have been reduced several times in recent years, although Congress has repeatedly delayed the reductions. This unpredictability makes it difficult for test developers to contemplate what the market for a particular test might look like, she said. In addition, the reduction in Medicare reimbursement threatens the stability of the clinical laboratory infrastructure, which is critical to the wellbeing of the country.
Other issues to consider when bringing a test to market include clinical guidelines and clinical/consumer uptake. Van Meter said that clinical guidelines often take years to be updated and thus may not reflect the most recent technologies; this can have a negative impact on accessibility and use of new tests. In addition, it can be difficult for clinicians to keep abreast of what diagnostic tests are available and how to employ them. There is a need, said Van Meter, to pursue new innovations in testing but also to leverage and improve the uptake of tests that are available today.
Discussion
Cost
One workshop participant observed that cost underpins many of the issues discussed during this session, including the cost of development, use of diagnostics, and reimbursement. They suggested that one roadblock may be reluctance on the part of hospitals to invest in new instrumentation and a reliance on outsourcing of testing to clinical laboratories. Whiteford agreed and added that outsourcing diagnostic tests also introduces transportation issues. He said that stakeholders should work together to figure out how to fund laboratory testing so that the potential of new technologies is realized. Van Meter emphasized that laboratory substructure—whether commercial labs, hospital labs, or point-of-care testing used in physician office labs—is part of the nation’s critical infrastructure. The recent reductions in Medicare reimbursement threaten the stability of this groundwork, and it is essential that the nation makes the necessary investments in building infrastructure to serve patients and communities, noted Van Meter. Technology is advancing to make new things possible but requires the infrastructure to support it.
Development and uptake
Workshop participants asked the panelists to comment on the role of different types of tests, specifically, lateral flow assays and multiplex tests for UTIs. Whiteford said that at-home tests that use lateral flow have been shown to be useful during COVID-19 for triaging patients. Daniel Bausch (Director of Emerging Threats & Global Health Security, FIND, The Global Alliance for Diagnostics) observed that many of the types of tests discussed thus far at the workshop are “near” point-of-care tests and that lateral flow assays can be used as “true” point-of-care tests in the hands of individuals at home. He added that there is an important role for these tests in the United States, but their utility is even more obvious in low- and middle-income countries, where infrastructure, electricity, and price can be barriers to testing in formal healthcare settings. Regarding multiplex tests, Van Meter said that such tests would be useful. For example, a patient presenting with respiratory symptoms could be efficiently tested for COVID-19, flu, and respiratory syncytial virus (RSV); however, it is unclear what the regulatory pathway for such a test would be, how its cost would be reimbursed, and what clinical guidelines would direct its use. Multiplex testing holds tremendous promise, but there is a need to clarify these issues to encourage development, she said. Jean Patel (Principal Scientific Affairs, Microbiology, Beckman Coulter Diagnostics) agreed that the issues of reimbursement and guidelines
influence test development and uptake. However, there are also examples of tests that are recommended for use and supported by reimbursement but are still not being widely used by clinicians (e.g., detecting colonization with carbapenem-resistant Enterobacterales [CRE] to prevent transmission in a healthcare setting). Van Meter agreed and suggested that there are mechanisms that can be leveraged to promote uptake in these circumstances. For example, public–private partnerships could fund a new technology add-on payment for tests that improve outcomes and enhance stewardship. Clinical guidelines can serve as substantial barriers to uptake and reimbursement, said Van Meter; new technologies often take years to make it into guidelines. She wondered aloud if there was a role for groups such as the Presidential Advisory Council on Combating Antibiotic-Resistant Bacteria (PACCARB) to highlight key areas and support efforts to develop recommendations that payers can use to make decisions.
Carroll observed that CRE testing is largely used for surveillance, rather than diagnosis, and suggested that one mechanism to promote uptake in this area is to include in budgets funds for CRE and other multidrug resistant (MDR) organism colonization for inpatient care units. Data on key MDR organisms are reported to the National Healthcare Safety Network, so the hospital has a vested interest in reducing indicator organisms. Carroll noted that using public health to leverage these types of AMR testing is one creative solution for getting tests paid for.
Another potential approach for improving development and uptake, said Burnam, is declaring a national health emergency. As seen during COVID-19, an emergency situation (and the associated regulatory flexibility) gives everyone “a big shot of espresso.” “Everything just starts working faster and better,” he said; investors look for opportunities to fund development and infrastructure, and stakeholders work together to find quick solutions. Declaring a national emergency for antimicrobial resistance could be the solution for many of the issues discussed today.
Bioethical considerations for clinical laboratories
Robin Patel (ID Physician, Clinical Microbiology Laboratory Director, Mayo Clinic) asked Cohen to comment on the bioethical considerations for clinical microbiology laboratory directors in the AMR space. Cohen responded that she would argue that directors do have bioethical duties and that a surveillance approach may be helpful when directors are considering what tests to bring in and how to offer them. For example, directors should be aware of what diseases are coming into their hospitals and what technologies are available for testing. Cohen said that physicians and lab directors “stand in the same kinds of shoes” in terms of their shared bioethical obligations to the patient to do no harm and to act for the good
of the patient. However, before they can do so, they need to be aware of what diagnostics are available. Cohen said there is a need to communicate better about new technologies and how they can improve clinical decision-making. Physicians in particular, she said, are overloaded with information and need support in staying up to date.
Coordination
A workshop participant observed that many speakers had mentioned the need for stakeholders to work together on guidelines for new diagnostics as well as issues around development, reimbursement, and delivery. They asked panelists to comment on the potential for a coordinating body to unite these efforts and to address issues simultaneously. R. Patel responded that developing guidelines can be a lengthy, intense process, in part because of the need for a body of published evidence. It is possible to develop guidelines quickly; for example, during the COVID-19 pandemic, the Infectious Diseases Society of America (IDSA) expeditiously put out guidelines. However, this required long hours of work and resources to facilitate the process. J. Patel noted that IDSA has published guidelines for treating infections caused by multi-drug resistant organisms; this can be an invaluable resource for clinicians and other stakeholders making decisions about what tests to provide.
Tradeoffs
A virtual workshop participant asked panelists how stakeholders could balance the tradeoffs between maximizing pathogen or resistance marker coverage versus practical point-of-care needs (i.e., tests that quickly inform clinical decisions). For example, should point-of-care tests broadly cover resistance markers, or should tests focus on common resistance markers? Carroll responded that priorities for testing will depend on the needs of the individual institution, the patient population, and the type of laboratory. However, it does not need to be an either/or situation. Carroll emphasized a need to simultaneously push point-of-care testing to inform treatment decisions and diagnostic tests that enable the collection of public health data for surveillance purposes.
Van Meter took a broader view of prioritization and said that efforts could be focused based on a variety of criteria: the type of bug, the areas where there is the most inappropriate prescribing, or care settings for which tools are available but are not being used. For example, she said, a study found that 30 percent of patients with upper respiratory symptoms who test positive for flu walk out of the doctor’s office with a prescription for antibiotics (Havers et al., 2014).
This page intentionally left blank.

















